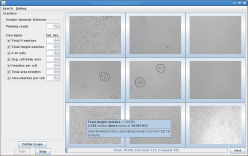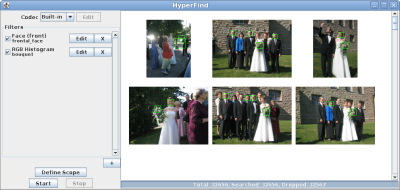
HyperFind is a sample domain-specific search application built on the OpenDiamond® platform. HyperFind enables users to quickly search through collections of unlabeled photographs (such as holiday photos). Our motivation is that digital cameras allow users to generate thousands of photos, yet few users have the patience to manually index them. Users typically want to find photos by content, but computers are not yet smart enough to understand the semantics of an image. HyperFind interactively filters images using a variety of pre-defined and user-trained filters. Partial results from earlier searches are used to refine subsequent queries allowing the user to iteratively refine the query as needed.
The current implementation of HyperFind supports user-defined color and visual texture filters that scan regions in every image. The user can create and refine these filters using image patches. HyperFind also supports popular shape-based filters such as face and pedestrian detectors (available in OpenCV). Many computer vision algorithms are well-suited for implementation on OpenDiamond since they can be treated as a cascade of filter stages. This enables much of the search to be downloaded to the storage node, where unpromising candidates can be quickly discarded.

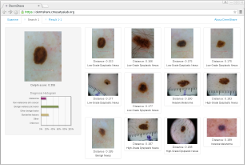
DermShare is an easy-to-use web-based tool to assist primary care physicians in more accurately detecting melanoma. The physician provides a dermatoscopic image of an unknown lesion, and the tool discovers and displays similar dermatoscopic images from a large library of samples, along with their pathology reports and diagnoses.
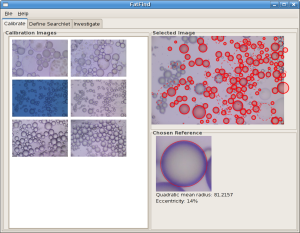
The task of manually counting adipocytes (fat cells) in cell microscopy images and characterizing their size is very time consuming. FatFind is a Diamond application, developed in collaboration with Merck Research, that exploits the almost perfectly circular shape of adipocytes in solution to efficiently locate fat cells.
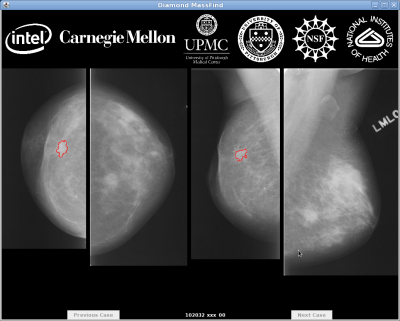
MassFind is an implementation of Interactive Search-Assisted Diagnosis (ISAD), specifically for mammography. ISAD is a collaborative research effort with the University of Pittsburgh and the University of Pittsburgh Medical Center.
The goal of ISAD is to enable doctors to make better decisions about a given case by providing a selection of similar annotated cases from a large database. For instance, a radiologist examining a suspicious mass could study labeled mammograms with similar conditions and weigh the outcome of their biopsy results before determining whether to recommend a biopsy. The fundamental challenge in developing ISAD systems is the identification of similar cases, not simply in terms of superficial image characteristics, but in a medically-relevant sense.
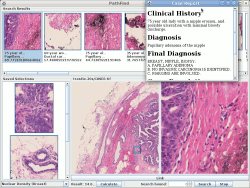
Based on analysis of expected workflow by a typical pathologist, a tool called PathFind has been developed. PathFind incorporates a vendor-neutral whole-slide image viewer that allows a pathologist to zoom and navigate a whole slide image just as he does with a microscope and glass slides today. The PathFind interface allows the pathologist to identify regions of interest on the slide at any magnification and then search for similar regions across multiple slide formats. The search results can be viewed and compared with the original image.
StrangeFind is an application for online anomaly detection across different modalities and types of data. It was developed with Merck to assist pharmaceutical researchers in automated cell microscopy, where very high volumes of cell imaging are typical. Anomaly detection is separated into two phases: a domain-specific image processing phase, and a domain-independent statistical phase. This split allows flexibility in the choice of image processing and cell type, while preserving the high-level aspects of the application. StrangeFind currently supports anomaly detection of adipocyte images (where the image processing analyzes sizes, shapes, and counts of fat cells), brightfield neurite images (where the image processing analyzes counts, lengths, and sizes of neurite cells), and XML files that contain image descriptors extracted by proprietary image processing tools.